RESEARCH - Henning Mootz
Protein chemistry / biotechnology
Posttranslational modifications of the Ubiquitin-type
Posttranslational modifications can regulate the activity of proteins and their interaction patterns with partner molecules in complex networks. It is one way how nature increases the structural and functional diversity of the proteome. A fundamental problem for the study of the role of a specific posttranslational modification, e.g. phosphorylation, lipidation, glycosylation, acetylation, or ubiquitination, is the oftentimes encountered difficulty or impossibility to obtain the modified protein in defined and pure form. This can be due to a number of reasons, for example, because of the different posttranslational modification machinery in the heterologous expression host, the low specificity of the respective transferase or ligase, or even our lacking knowledge of the modifying enzyme. A chemical approach to site-specifically install the posttranslational modification can circumvent these problems and lead to the preparation of defined proteins for biochemical, structural, or cell biological studies. It may also provide access to analogs of the posttranslational modification that are useful to understand their structure-function relationship or to artificial conjugates that are stable against enzymatic cleavage.
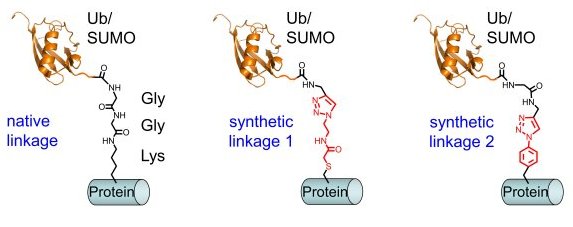
We are interested in the posttranslational modification of proteins with ubiquitin (Ub) and ubiquitin-like (Ubl) proteins, such as SUMO (small ubiquitin-like modifier). These proteins are typically linked via an isopeptide bond involving a lysine residue side chain of the target protein and their own carboxy terminus. The 76 amino acid ubiquitin is best known for its role in the ubiquitin-proteasome pathway for protein degradation by Lys48-linked poly-ubiquitinylation, however, mono-ubiquitinylation and poly-ubiquitinylation on different lysine residues also serve regulatory functions in many biological processes, e.g. signalling and endocytosis. SUMOylation is highly dynamic and involved in, for example, nuclear transport and DNA-repair.
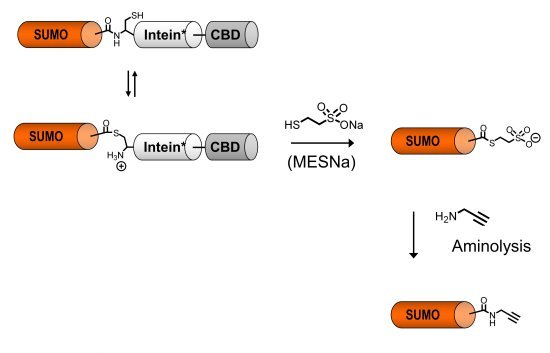
To prepare site-specific protein-ubiquitin and protein-SUMO conjugates we have devised a chemical approach based on Cu(I)-catalyzed click chemistry (1-4). This bioorthogonal cycloaddition between an azide and an alkyne yields a stable triazole linkage and can be performed in the presence of other functional groups present in proteins. Importantly, this linkage is stable towards ubiquitin and SUMO-specific isopeptidases that catalyze hydrolysis of the native isopeptide bond. We use these synthetic conjugates for biochemical studies and for the identification of new binding partners that recognize the new composite interface created by the posttranslational protein modifier.
Selected references
Weikart, N. D., and Mootz, H. D. (2010)
Generation of site-specific and enzymatically stable conjugates of recombinant proteins with ubiquitin-like modifiers by the Cu(I)-catalyzed azide-alkyne cycloaddition, Chembiochem 11, 774-777.
Sommer, S., Weikart, N. D., Brockmeyer, A., Janning, P., and Mootz, H. D. (2011)
Expanded click conjugation of recombinant proteins with ubiquitin-like modifiers reveals altered substrate preference of SUMO2-modified Ubc9, Angew Chem Int Ed 50, 9888-9892.
Weikart, N. D., Sommer, S., and Mootz, H. D. (2011)
Click synthesis of ubiquitin dimer analogs to interrogate linkage-specific UBA domain binding, Chem Commun 48, 296-298.
Sommer, S., Ritterhoff, T., Melchior, F., and Mootz, H. D. (2015)
A stable chemical SUMO1-Ubc9 conjugate specifically binds as a thioester mimic to the RanBP2-E3 ligase complex, Chembiochem 16, 1183-1189.

