Demonstration
Proteomatic demonstration
Before you continue, please download Proteomatic. After that, download the following package which contains all necessary files for a quick demonstration:
| Proteomatic demonstration package | |
|---|---|
 |
proteomatic-demonstration.zip (73.9 MiB) |
The demonstration package above contains the following files:
- spectra: ~600 full scans and ~1,400 fragmentation scans from a LC-MS/MS measurement of a 14N/15N metabolically labeled sample of isolated Chlamydomonas reinhardtii chloroplasts grown under different conditions assessing the iron deficiency response of the alga. All spectra are combined within a single file.
- protein databases: gene models deduced from the nuclear, chloroplast, and mitochondrial genomic sequences of Chlamydomonas reinhardtii.
- pipeline: a Proteomatic pipeline including:
- protein identification at a user-defined false positive rate using OMSSA
- protein quantitation using qTrace
Please download the file and unpack it. Now start Proteomatic and load the Proteomatic Demonstration.pipeline file via drag and drop or by choosing Pipeline → Open pipeline. After downloading and unpacking the necessary software packages, the following pipeline will be loaded:
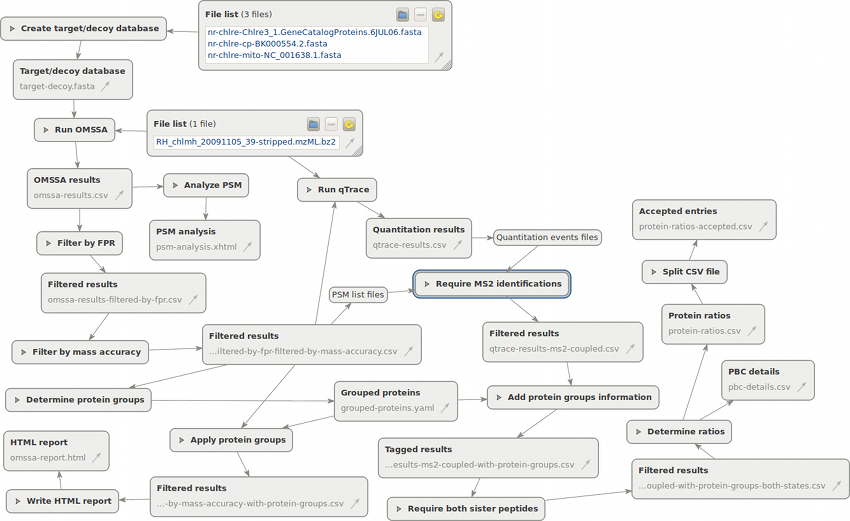
Handling Proteomatic is designed to be quite intuitive (the most important points being Ctrl+scroll wheel for zooming and the Show all button). Please have a look at the getting started page for a quick tutorial.
Now run the pipeline by clicking Start. Downloading external programs and running the pipeline should not take more than 10 minutes. All result files will be written into the results directory in the package, so that no traces remain when you delete the extracted package.