Research Interests of the
Biotic Plant Interactions Group



Parasitic plants of the broomrape family (Orobanchaceae) steal both water and nutrients from other plants. Amongst the world's most problematic parasitic weeds are broomrapes (Orobanche s.l.) and witchweeds (Striga), which attack the roots of important cereals and vegetable crops. Fundamental knowledge of the molecular evolutionary forces and constraints influencing the evolution of parasitism within Orobanchaceae would help us to develop more effective and sustainable pest control procedures.
The goal of this research is to identify commonalities as well as adaptive and nonadaptive genetic reconfigurations that are associated with the transition from an autotrophic to a holoparasitic way of life within the Orobanchaceae. The proposed research program exploits the unique, natural genetic diversity of the broomrape family, which comprises the full trophic spectrum including nonparasites, hemiparasites of various specializations and holoparasites that have evolved multiple times independently. We will test whether and which genetic changes precede or follow the transition to holoparasitism and if these reconfigurations occur in a predictable order.
A primary aim is to disentangle the nature and direction of molecular evolutionary shifts in the genes of the parasites. To this end, we use qualitative and quantitative RNA and DNA sequencing to compare the gene sets and gene expression patterns between several autotrophic and heterotrophic Orobanchaceae. A key aspect lies in employing existing and devising novel probabilistic models to investigate our research hypotheses in a unified computational framework that fuses genotypic and phenotypic information. This study thus will allow assessing the interplay between changes in the genetic profile of closely related parasitic plants and the underlying molecular evolutionary forces that act as fuel for macroevolutionary change - and vice versa.
The well-developed Orobanchaceae model system in combination with state-of-the-art molecular evolutionary methods provides the ideal tool to develop an explanatory model for the genetic changes associated with trophic specialization in plants. The more than five independent transitions to holoparasitism in the family will enable us to specifically examine whether adaption to this way of life has occurred in at least as many different ways, and help us to identify the functions associated with these lifestyle. Our research objectives thereby focus on key questions concerning the genetics of Orobanchaceae and, at the same time, address unresolved issues of molecular rate variation in plants. Thus, our work will contribute towards unraveling the causes and consequences of the transition to parasitism in plants and identifying its underlying mechanisms. Comparative biocomputational approaches in evolutionary biology will further the development of a generalized explanatory model of the effects of genomic and phenotypic shifts, which will impact research areas in many branches of the life sciences.
This project is mainly funded by the German Research Foundation (Deutsche Forschungsgemeinschaft) in the Emmy Noether-Program.


Parasitism represents the most extreme interaction between plants, where the parasitic plant steals water and nutrients from another plant. The dependency on a host represents an isolating barrier absent from nonparasites, which eventually changes diversification patterns as parasitism intensifies. The Orobanchaceae family represents a natural model system for studying host-related diversification in plants. The species-rich family comprises nonparasitic species as well as hundreds of parasites that differ in their lifestyle as photosynthetic or nonphotosynthetic parasites, their degree of host dependency, and their host preferences. By attacking the roots of important crops such as maize, legumes, canola, and many vegetables including tomato and potato, some Orobanchaceae such as Striga, Orobanche, and Phelipanche have become the agro-economically most important parasitic weeds. The potential for the evolution of more agricultural pests is high, because many wild Orobanchaceae share similar ecological preferences and/or life histories with their noxious relatives. Despite this, we lack phylogenetic models for the spatio-temporal, host-ecological, and genomic evolution of the entire family, which would allow assessing the risk of co-domesticating new virulent ecotypes.
The goal of this research is to understand the mechanisms and tempo of the diversification of parasitic Orobanchaceae, for which we will reconstruct the biogeographical and host-ecological history of Orobanchaceae, test if host or abiotic environmental preferences, or both, have drive their diversification, and analyze the evolution of gene diversity en route to a fully parasitic lifestyle. Molecular evolutionary and phylogenetic-comparative methods provide the sound basis for testing spatio-temporal, evolutionary-ecological, and functional-genetic hypotheses. We aim to disentangle the host-effect from abiotic environmental influences that contribute to parasite diversification by testing in a Bayesian framework, if shifts of diversification rates in relation to changes in the degree of trophic specialization correlate with shifts of host ranges and/or environmental preferences. Genomic data from parasites of varying trophic levels and host preferences will allow us to isolate those genes that are potentially involved in parasitic processes in general, and nutritional specialization in particular. Because of the arms race with their hosts, “parasitism genes” evolve under distinct selectional regimes, which we will detect with a rigorous analysis of molecular evolutionary rates and selection pressures in a Maximum Likelihood and Bayesian framework. In the long run, the fundamental insights into the biology of parasitic Orobanchaceae to be delivered by this project will contribute to the international efforts of managing infestations of parasitic weeds in agriculture.
This project is supported by the German-Israeli Foundation for Scientific Research and Development in its Young PI-Program.


The Broomrape family (Orobanchaceae) contains the worst agricultural weeds. While we know that many obligate parasites of Orobanchaceae rely on host-exuded molecules for their earliest development, it remains unknown how far lifecycle synchronization extends and whether it includes reproduction. This project analyzes the molecular evolution and function of gene complexes involved in flowering time regulation of Orobanchaceae. We will evaluate whether a shift from annual to perennial hosts, or vice versa, alters the life history preferences of the parasites. On the other hand, our experiments test whether the induction of flowering is an autonomous process in holoparasitic plants.
To this end, we carry out molecular evolutionary analyses to assess the selectional footprints in the parasites' flowering time regulators in relation to their life history preferences. Using a custom bioinformatic pipeline, we extract the sets of functionally validated flowering genes from our genome and transcriptome data. The resulting gene and protein alignments are then subjected to molecular evolutionary analysis, for which we use branch-site random effects and trait-rate fusion models under the maximum likelihood paradigm.
Further, we use these data to design parasite-specific RNA-silencing constructs, which allows us to target different flowering pathways of the broomrape species Phelipanche ramosa and P. aegyptiaca via host induced gene-silencing (HIGS). Following a successful transformation of host plants like Arabidopsis, tomato, and tobacco, we monitor phenotypic changes in parasite flowering through rhizotron and pot assays.
Our combination of molecular evolutionary analyses of genes sets from a broad range of parasites and functional analysis promise to uncover the genetic mechanisms underlying the control and induction of parasitic plant flowering. This knowledge is likely to bring forward novel targets for parasitic weed management. Preventing the parasite from reproducing could help controlling its seed bank whenever an infestation was unavoidable.
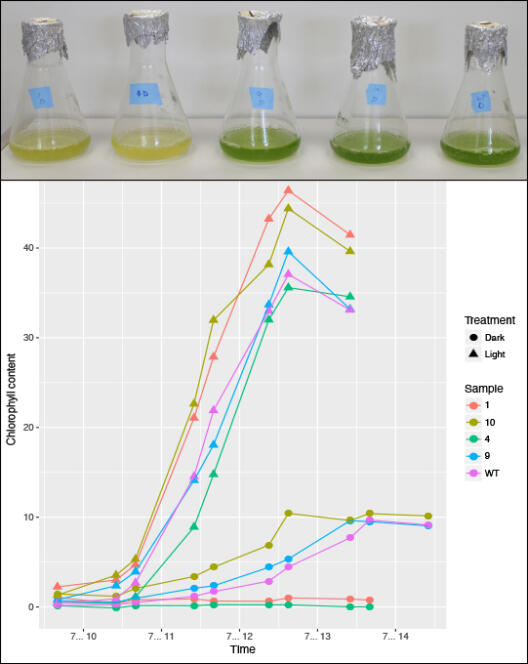
The chlorophytic green alga Chlamydomonas reinhardtii is not limited to a photo-autotrophic lifestyle but can also live heterotrophically under no-light conditions in the presence of acetate as an external carbon source. This form of myxotrophy can be regarded as a major eco-physiological and evolutionary advantage. It allows C. reinhardtii and many other algae to adapt rapidly to natural fluctuations of the available light and nutrient resources. Niche shifting to a permanent heterotrophic lifestyle will evolve highly adapted C. reinhardtii populations, whose specialist individuals exhibit an increased fitness under zero-light conditions compared to the original myxotrophs that experience no-light environments only temporarily. The function of genes and protein complexes conveying photo-autotrophy will be relaxed of purifying selection in the heterotrophic selection line, because genes for photosynthesis are of no more use. An evolutionary experiment that controls for various selection pressures will aim to elucidate the timing and series of adaptive genomic and metabolic changes associated with the transition from mixotrophy to permanent heterotrophic conditions, thus simulating the transition from facultative to obligate heterotrophy in plants. Specifically, we will investigate”
To this end, we will evolve three lines of C. reinhardtii for >1,500 generations under heterotrophic (zero-light with acetate), mixotrophic (light plus acetate), and photo-autotrophic conditions (light without external carbon source). We will sample from up to ten replicates of each selection line per strain every 100th generation to measure biomass production, the assembly efficiency of the photosynthesis machinery, the metabolization of metals such as iron and magnesium, and the mating success between the various selection lines and their ancestors. We expect to observe an early fitness effect in the growth rates after less than 600 generations, with a fixation and accumulation of genomic and metabolic differentiations being highest in the heterotrophs. Every 200th generation, we will perform phenotypic, genomic, transcriptomic, and proteomic assays to infer the genetic responses to light exposure in the absence of external carbon. These data will be compared to the reference phenotypes, genomes, transcriptomes, and proteomes of the corresponding starter lines. Besides shedding light on the early functional reconfigurations during the transition to a secondary heterotrophic lifestyle, this project will generate a wealth of naturally selected C. reinhardtii lines with fine-tuned adaptations to various growth conditions that may aid future research in plant evolutionary biology as well as biotechnological application such as the production of biofuels under no-light conditions.
This research program aims to develop an integrated diagnostics procedure based on DNA (meta)barcodes extracted from genomic resources of wild and weedy parasites throughout the biodiversity range of Orobanchaceae. The envisioned workflow combines diagnostics and evolutionary inferences to identify parasites to their lowest taxonomic levels in a user-friendly toolkit. Sets of barcodes that qualify as ecotype-sensitive identifiers will be prepared for practical, semi-quantitative pull-down assays. Besides testing the discriminative power and sensitivity of suitable barcode combinations in silico, the integration of evolutionary analyses and, thus, the placement of marker development in an explicit phylogenetic framework, will improve our ability to delimitate taxa in rapidly evolving lineages where cryptic diversity is prevalent. The envisioned toolkit for high-throughput identification of parasitic weeds directly from environmental samples will allow assessing the genetic diversity of parasite seed banks and monitoring soil infestation levels over time while needing to convince by practicability and sensitivity experimentally. The development of a diagnostic workflow for Orobanchaceae will generate a positive feedback loop between basic eco-evolutionary research and applied life science en route to an ecotype-specific control of parasitic weeds, and potentially other common plant pests or diseases.
We are looking for enthusiastic (under)graduate students or post-docs to help us challenge early version of our diagnostics tool – experimentally and/or bioinformatically! Interested researchers please email us, or drop by during regular office hours on Tuesdays and Thursdays.

Biophysical carbon concentrating mechanisms (CCMs) operating at the single-cell level have evolved independently in some eukaryotic algae and a single lineage of land plants, the hornworts. An essential component for an efficient biophysical CCM is a pyrenoid, which represents a specialized compartment inside chloroplasts that mainly comprise the CO2-fixing enzyme RuBisCO. Hornworts with pyrenoids fix significantly more carbon than their relatives without pyrenoids. Given the repeated gains and losses of pyrenoids in hornworts during the last 50 million years, we may assume that their assembly is potentially controlled by a few master regulators of eco-evolutionary relevance. In a joint effort, we will combine comparative -omics with reverse genetics tools to study the genetics, function, and molecular basis of pyrenoid-based CCM in hornwort plastids under different environmental conditions. Guided by ultrastructure-based monitoring of the pyrenoid assembly in hornworts, we aim to identify the genetic toolkit of biophysical CCM in hornworts through two interdependent approaches: First, we aim to predict candidate CCM components in silico though a set of homology searches that compare the hornwort gene set with algal CCM component. Second, we employ an exploratory gene and protein (co)expression profiling of isolated plastids collected under low vs. high CO2 concentrations and under flooding. A strength of our experimental design is that we contrast pairs of pyrenoid bearing and pyrenoid lacking hornwort species and consider the unusual association of the hornworts with cyanobacterial symbionts. This exploratory analysis is necessary because there is no guarantee that CCM induction and biophysical function of pyrenoids relies only on a set of homologous genes in hornworts and algae. Finally, we will investigate pyrenoid functionality under various environmental conditions and regarding its role for the cyanobacterial association. Specifically, we aim to conduct localization and functional validation analyses for a core set of genes discovered in our CCM gene prediction approaches. These experiments are possible through our recent advances to establish Anthoceros agrestis and other hornwort species as a tractable model system. Together, our collaborative project will not only allow a comparison of the mechanisms of pyrenoid assembly between algae and hornworts, but also reveal general principles and species-specific innovations in the evolution of carbon-concentrating plastids. Above that, focusing on and understanding the basis of land plant CCM instead of only the algal form could eventually contribute to efficiently engineer pyrenoid assembly and boost photosynthetic efficiency of crops.